Almost a decade ago, UO graduate student Jennifer Hampton Hill made a fortuitous find: A protein made by gut bacteria that triggered insulin-producing cells to replicate. The protein was an important clue to the biological basis for Type 1 diabetes.
Hill has continued exploring the protein, called BefA, as a postdoctoral researcher at the University of Utah. And biology professor Karen Guillemin’s lab at the UO has kept studying BefA, too. Alongside other colleagues, they’ve now uncovered new insights into what BefA does and why bacteria make it.
Those discoveries have “important, profound implications,” said Guillemin, whose lab in the biology department is part of the College of Arts and Sciences. “If we understand how BefA works, it could give us a way to stimulate beta cell production therapeutically.”

The body needs insulin to regulate blood sugar, but insulin is only made by a select type of cells in the pancreas called beta cells. And there’s a narrow window of time during early childhood development when beta cells replicate and expand their population. In people with Type 1 diabetes, the immune system attacks beta cells and depletes their population, limiting insulin production.
Microbiome stimulation of immune development helps properly educate the immune system and prevent autoimmunity. The work by Guillemin’s team suggests an additional role for the microbiome: It stimulates growth of the beta cell population early in development, buffering against later depletion by autoimmune attack.
Beta cell population growth “is happening at the same time that microbial communities are diversifying in the gut,” Hill said. “A hallmark of diabetes is kids who develop it tend to have a less diverse gut microbiome. It’s possible they're missing some of the bacteria that make BefA.”
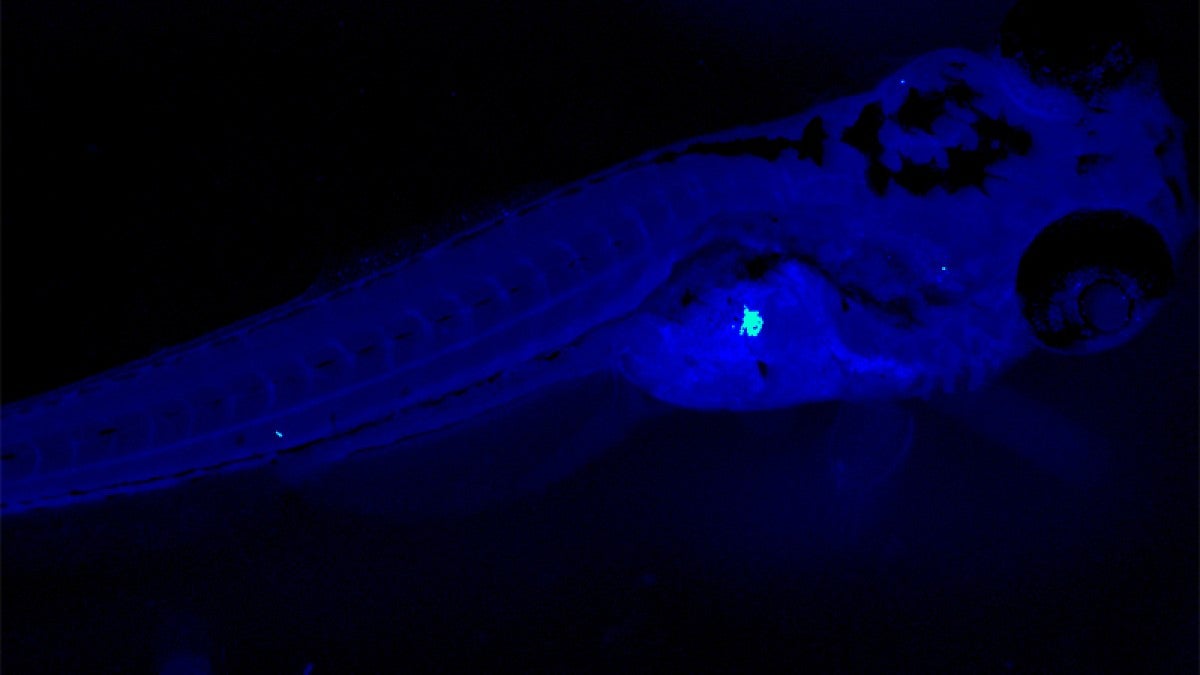
In their most recent paper, Hill, Guillemin and their colleagues take a deeper look at BefA. They captured detailed images of BefA’s structure to identify the parts of it that interact with cell membranes. Then, through a series of experiments in zebrafish, mice and cultured cells, the researchers sketched a picture of BefA’s function.
BefA can disrupt the membranes of many kinds of cells, both bacterial and animal, they showed. It makes sense that gut bacteria would attack competing bacteria. But unexpectedly, they also found that BefA’s attacks on the membranes of insulin-producing cells triggered those cells to reproduce.
The finding suggests that bacterial warfare in the gut can have collateral beneficial effects on the body, boosting the population of cells that can make insulin throughout the lifespan.
The team also tested a mutated version of BefA that was modified so that it couldn’t mess with cell membranes. That version of the protein didn’t impact beta cell production, further suggesting that membrane damage is driving BefA’s effects.
“There are other examples in developmental biology where poking holes in membranes is critical in stimulating development,” Hill said, but the researchers don’t yet know exactly how the damage is triggering cell replication here.
And they don’t know why BefA, which can actually alter the membranes of many kinds of cells, targets beta cells so specifically.
“We think that there's something special about beta cells that they may be highly sensitized to respond to cues that cause membrane permeabilization,” Hill said. “They're the only cell type in the whole body that can secrete insulin; they're highly important.”
Hill was awarded the NOSTER & Science Microbiome Prize this year for her work on BefA. The annual award is given to an early-career scientist who has contributed new understanding to microbiome research that could influence human health.
“The microbiome plays a role in educating the immune system. If you don't have that education, the immune system can be hyper-reactive,” Guillemin said. “We think there's also this other layer here. If you don't develop a pool of beta cells against future disruption, you're more at risk for Type 1 diabetes.”
And a healthy, diverse microbiome plays a key role in building that cell population.
In the future, Guillemin’s team imagines possible therapeutic applications for the finding. For example, proactively fortifying the microbiomes of high-risk infants with BefA-producing bacteria could prevent them from later developing Type 1 diabetes.
—By Laurel Hamers, University Communications